- Published on
Gaussian Naive Bayes
- Authors

- Name
- Mai Khoi TIEU
- @tieukhoimai
Table of Contents
Gaussian Naive Bayes
Disclaimer: This article is based on my personal lecture notes from my studies at Machine Learning for Data Science 2023/2024 course: Bayesian Learning. The content reflects my understanding and interpretation of the course material.
Objective
Given a data point x, what is the probability of x belonging to some class c?
We have C class and need to find which group of , instead of finding exactly the label, given a data point x, we find the probability of x belonging to some class c or find or
We define the class of each point by choosing the class c with the highest probability
Bayes’ theorem
Naive Bayes Classifier formula
or
where
- consists with with = observation and with is the number of features
- is Posterior Probability
- is Likelihood of feature given that their class is T
- is Prior Probability
- is Marginal Probability or called Evidence
We have
Implementation
We choose Iris as a dataset.
from sklearn.datasets import load_iris
import matplotlib.pyplot as plt
x, y = load_iris( return_X_y=True )
There are
- 3 classes
- 150 observations with 4 features
Likelihood of each feature per class
Computing depends on the type of data. There are three commonly used types: Gaussian Naive Bayes, Multinomial Naive Bayes, and Bernoulli Naive Baye
We will consider the Gaussian distribution, with two common distributions: the univariate normal distribution (or Gaussian density) and the multivariate normal distribution (or multivariate density).
Univariate normal distribution or Gaussian density
This distribution is described by two parameters: the mean and the variance .
The value of can be any real number, indicating the location of the peak, where the probability density function reaches its highest value.
Example: Plot the gaussian distributions (likelihood) with the < and same variance
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import norm
# Generate the functions' domain
x = np.linspace(0,20,100)
# Define two gaussian distributions
p1 = norm.pdf(x, 2.5, 1.5)
p2 = norm.pdf(x, 5.5, 1.5)
# Plot the gaussian distributions (likelihood)
plt.plot( x, p1, 'b')
plt.plot( x, p2, 'g')
plt.title('Likelihood distributions')
plt.ylabel('$p(x|c)$')
plt.xlabel('$x$')
plt.show()
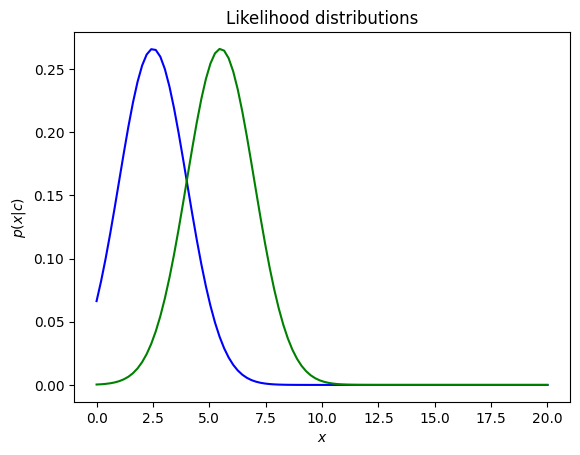
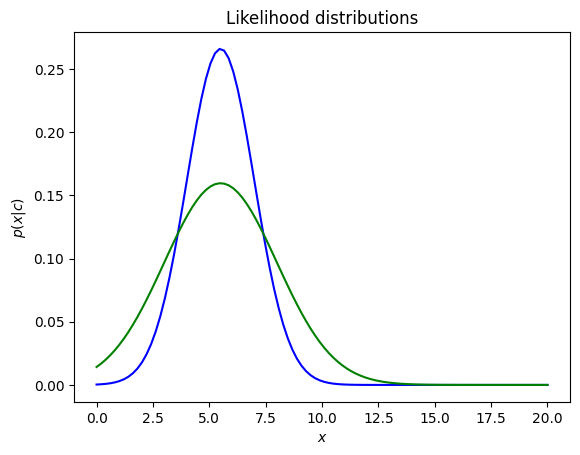
The value of is a positive value, with representing the spread of this distribution. A large indicates a wide range of output values, and vice versa.
Example: Plot the gaussian distributions (likelihood) with the < and same variance
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import norm
# Generate the functions' domain
x = np.linspace(0,20,100)
# Define two gaussian distributions
p1 = norm.pdf(x, 5.5, 1.5)
p2 = norm.pdf(x, 5.5, 2.5)
# Plot the gaussian distributions (likelihood)
plt.plot( x, p1, 'b')
plt.plot( x, p2, 'g')
plt.title('Likelihood distributions')
plt.ylabel('$p(x|c)$')
plt.xlabel('$x$')
plt.show()
Multivariate normal distribution (or multivariate density)
This is the general case of the normal distribution when the variable is multidimensional, assuming it has dimensions.
where
- is -component column vector
- is -component mean vector
- is matrix and a positive definite symmetric matrix.
Back to our example with Iris dataset, we compute the likelihood for each sample and each class.
import numpy as np
from scipy.stats import multivariate_normal
# Identify members of each class -> 4 column vector with 150 obs
cl1 = x[ y==0, :]
cl2 = x[ y==1, :]
cl3 = x[ y==2, :]
# Compute the mean (centroid) of each class -> return a 4 row vector
m1 = np.mean(cl1, axis = 0)
m2 = np.mean(cl2, axis = 0)
m3 = np.mean(cl3, axis = 0)
# Compute the covariance matrix for each class -> return a 4x4 matrix (4 features)
C1 = np.cov(cl1.T) # or using C1 = np.cov(cl1, rowvar=False)
C2 = np.cov(cl2.T)
C3 = np.cov(cl3.T)
# compute the likelihood for each sample and each class
lik1 = multivariate_normal.pdf(x, mean=m1, cov=C1)
lik2 = multivariate_normal.pdf(x, mean=m2, cov=C2)
lik3 = multivariate_normal.pdf(x, mean=m3, cov=C3)
Prior Probability
Each value , where , can be determined as the frequency of occurrence of class in the training data.
# Define the priors
P_c1 = np.size(cl1)/np.size(x) # 1/3
P_c2 = np.size(cl2)/np.size(x) # 1/3
P_c3 = np.size(cl3)/np.size(x) # 1/3
Marginal distribution or Evidence
We compute in this case of 3 classes
p_x = (lik1 * P_c1) + (lik2 * P_c1) + (lik3 * P_c1)
Posterior Probabilities
# Compute posterior probabilities
post1 = (lik1 * P_c1) / p_x # shape = (150,)
post2 = (lik2 * P_c2) / p_x # shape = (150,)
post3 = (lik3 * P_c3) / p_x # shape = (150,)
post = np.vstack((post1,post2,post3)) # shape = (3,150)
Making Prediction
When is large and the probabilities are small, the expression on the right-hand side of equation above becomes a very small number, leading to potential numerical inaccuracies during computation. To address this, equation above is often rewritten in an equivalent form by taking the logarithm of the right-hand side
In this case, with , the results of the logarithm of the posterior probabilities and the posterior probabilities are equal.
logpost = np.log(post)
prediction = np.argmax(logpost,axis=0)
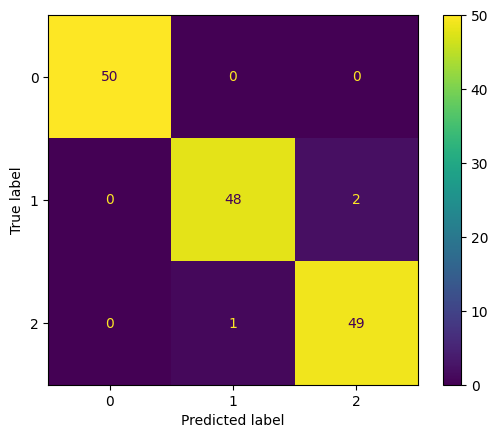
accuracy = np.sum(prediction == y)/len(y)
# Classifier accuracy: 98.00%
print('Classifier accuracy: ' + "{0:.2f}".format(accuracy*100) + '%')
# Visualize confusion matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y,prediction)
from sklearn.metrics import ConfusionMatrixDisplay
cm_display = ConfusionMatrixDisplay(cm).plot()
Reference
- Duda, R. O., Hart, P. E., & Stork, D. G. (2001). Chapter 2: Bayesian decision theory. In Pattern Classification. Wiley.
- Naive Bayes Classifier Implementation. GitHub.
- Chapter 11: Naive Bayes Classifier. Github
- Understanding by Implementing: Gaussian Naive Bayes. Towards Data Science.